Note
Click here to download the full example code
Spurious PAC with spikes¶
This example demonstrates how a spike train can generate a significant amount of phase-amplitude coupling (PAC), even though there is no nested oscillations. This phenomenon is usually referred as spurious PAC.
References¶
Dupre la Tour et al. (2017). Non-linear Auto-Regressive Models for Cross-Frequency Coupling in Neural Time Series. bioRxiv, 159731.
import numpy as np
import matplotlib.pyplot as plt
import scipy.signal
from pactools import Comodulogram, REFERENCES
from pactools.utils.pink_noise import pink_noise
from pactools.utils.validation import check_random_state
def simulate_spurious_pac(n_points, fs, spike_amp=1.5, spike_fwhm=0.01,
spike_fq=10., spike_interval_jitter=0.2,
random_state=None):
"""Simulate some spurious phase-amplitude coupling (PAC) with spikes
References
----------
Gerber, E. M., Sadeh, B., Ward, A., Knight, R. T., & Deouell, L. Y. (2016).
Non-sinusoidal activity can produce cross-frequency coupling in cortical
signals in the absence of functional interaction between neural sources.
PloS one, 11(12), e0167351
"""
n_points = int(n_points)
fs = float(fs)
rng = check_random_state(random_state)
# draw the position of the spikes
interval_min = 1. / float(spike_fq) * (1. - spike_interval_jitter)
interval_max = 1. / float(spike_fq) * (1. + spike_interval_jitter)
n_spikes_max = np.int(n_points / fs / interval_min)
spike_intervals = rng.uniform(low=interval_min, high=interval_max,
size=n_spikes_max)
spike_positions = np.cumsum(np.int_(spike_intervals * fs))
spike_positions = spike_positions[spike_positions < n_points]
# build the spike time series, using a convolution
spikes = np.zeros(n_points)
spikes[spike_positions] = spike_amp
# full width at half maximum to standard deviation convertion
spike_std = spike_fwhm / (2 * np.sqrt(2 * np.log(2)))
spike_shape = scipy.signal.gaussian(M=np.int(spike_std * fs * 10),
std=spike_std * fs)
spikes = scipy.signal.fftconvolve(spikes, spike_shape, mode='same')
noise = pink_noise(n_points, slope=1., random_state=random_state)
return spikes + noise, spikes
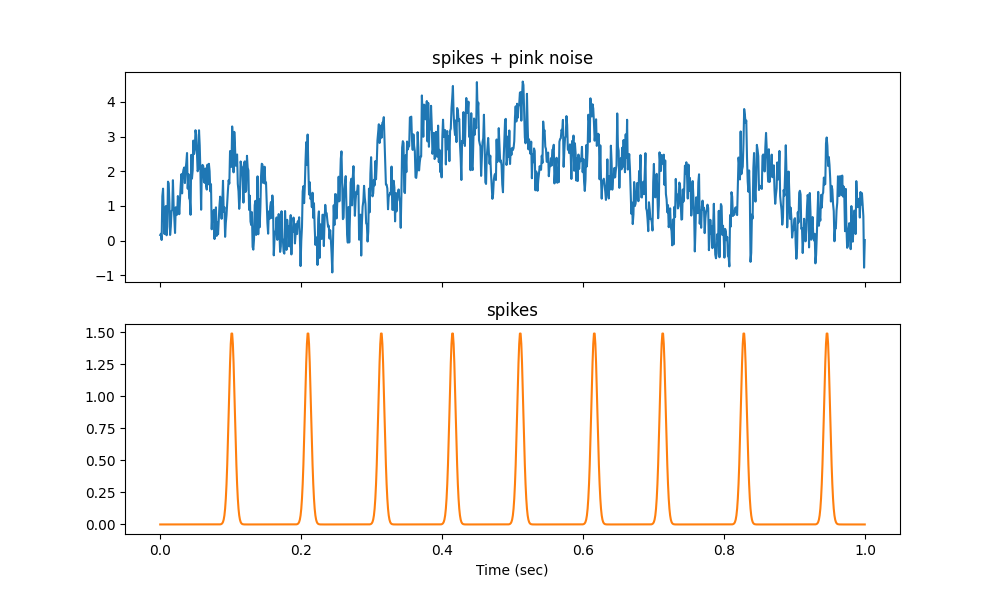
Let’s generate the spurious PAC signal
fs = 1000. # Hz
n_points = 60000
spike_amp = 1.5
random_state = 0
# generate the signal
signal, spikes = simulate_spurious_pac(
n_points=n_points, fs=fs, random_state=random_state, spike_amp=spike_amp)
# plot the signal and the spikes
n_points_plot = np.int(1. * fs)
time = np.arange(n_points_plot) / fs
fig, axs = plt.subplots(nrows=2, figsize=(10, 6), sharex=True)
axs[0].plot(time, signal[:n_points_plot], color='C0')
axs[0].set(title='spikes + pink noise')
axs[1].plot(time, spikes[:n_points_plot], color='C1')
axs[1].set(xlabel='Time (sec)', title='spikes')
plt.show()
Out:
/home/tom/work/github/pactools/examples/plot_spurious_pac.py:82: UserWarning: Matplotlib is currently using agg, which is a non-GUI backend, so cannot show the figure.
plt.show()
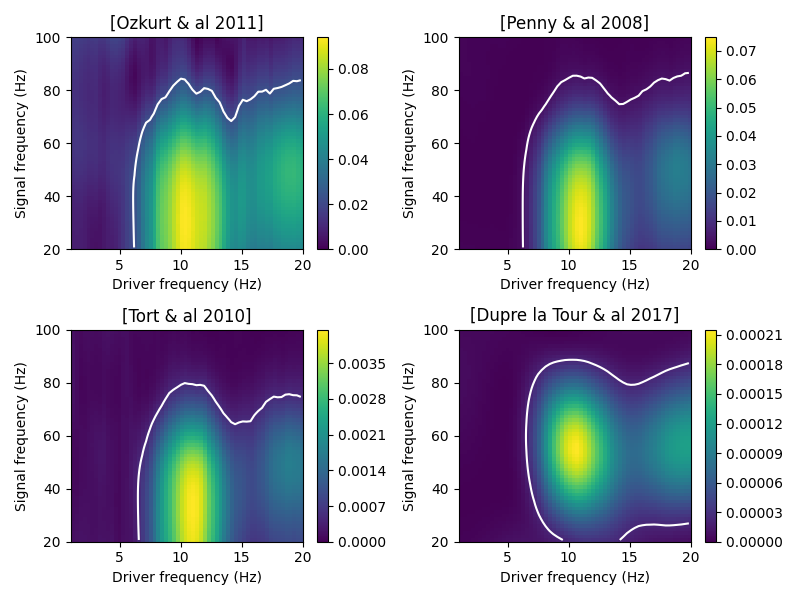
Compute the comodulograms
methods = ['ozkurt', 'penny', 'tort', 'duprelatour']
low_fq_range = np.linspace(1., 20., 60)
high_fq_range = np.linspace(low_fq_range[-1], 100., 60)
low_fq_width = 4. # Hz
# A good rule of thumb is n_surrogates = 10 / p_value. Example: 10 / 0.05 = 200
# Here we use 10 to be fast
n_surrogates = 10
p_value = 0.05
n_jobs = 11
# prepare the plot axes
n_lines = 2
n_columns = int(np.ceil(len(methods) / float(n_lines)))
figsize = (4 * n_columns, 3 * n_lines)
fig, axs = plt.subplots(nrows=n_lines, ncols=n_columns, figsize=figsize)
axs = axs.ravel()
for ax, method in zip(axs, methods):
# compute the comodulograms
estimator = Comodulogram(fs=fs, low_fq_range=low_fq_range,
high_fq_range=high_fq_range,
low_fq_width=low_fq_width, method=method,
n_surrogates=n_surrogates, n_jobs=n_jobs)
estimator.fit(signal)
# plot the comodulogram with contour levels
estimator.plot(contour_method='comod_max', contour_level=p_value, axs=[ax],
titles=[REFERENCES[method]])
fig.tight_layout()
plt.show()
Out:
[ ] 0% | 0.00 sec | comodulogram: ozkurt
[ ] 2% | 7.11 sec | comodulogram: ozkurt
[. ] 3% | 7.86 sec | comodulogram: ozkurt
[.. ] 5% | 8.70 sec | comodulogram: ozkurt
[.. ] 7% | 9.36 sec | comodulogram: ozkurt
[... ] 8% | 10.14 sec | comodulogram: ozkurt
[.... ] 10% | 10.90 sec | comodulogram: ozkurt
[.... ] 12% | 11.58 sec | comodulogram: ozkurt
[..... ] 13% | 12.24 sec | comodulogram: ozkurt
[...... ] 15% | 12.78 sec | comodulogram: ozkurt
[...... ] 17% | 13.39 sec | comodulogram: ozkurt
[....... ] 18% | 13.89 sec | comodulogram: ozkurt
[........ ] 20% | 14.51 sec | comodulogram: ozkurt
[........ ] 22% | 15.04 sec | comodulogram: ozkurt
[......... ] 23% | 15.76 sec | comodulogram: ozkurt
[.......... ] 25% | 16.50 sec | comodulogram: ozkurt
[.......... ] 27% | 17.15 sec | comodulogram: ozkurt
[........... ] 28% | 17.78 sec | comodulogram: ozkurt
[............ ] 30% | 18.27 sec | comodulogram: ozkurt
[............ ] 32% | 18.89 sec | comodulogram: ozkurt
[............. ] 33% | 19.82 sec | comodulogram: ozkurt
[.............. ] 35% | 20.69 sec | comodulogram: ozkurt
[.............. ] 37% | 21.52 sec | comodulogram: ozkurt
[............... ] 38% | 22.49 sec | comodulogram: ozkurt
[................ ] 40% | 23.41 sec | comodulogram: ozkurt
[................ ] 42% | 23.98 sec | comodulogram: ozkurt
[................. ] 43% | 24.44 sec | comodulogram: ozkurt
[.................. ] 45% | 25.17 sec | comodulogram: ozkurt
[.................. ] 47% | 26.03 sec | comodulogram: ozkurt
[................... ] 48% | 26.59 sec | comodulogram: ozkurt
[.................... ] 50% | 27.45 sec | comodulogram: ozkurt
[.................... ] 52% | 28.73 sec | comodulogram: ozkurt
[..................... ] 53% | 29.47 sec | comodulogram: ozkurt
[...................... ] 55% | 30.71 sec | comodulogram: ozkurt
[...................... ] 57% | 31.87 sec | comodulogram: ozkurt
[....................... ] 58% | 32.54 sec | comodulogram: ozkurt
[........................ ] 60% | 33.33 sec | comodulogram: ozkurt
[........................ ] 62% | 33.69 sec | comodulogram: ozkurt
[......................... ] 63% | 34.09 sec | comodulogram: ozkurt
[.......................... ] 65% | 34.51 sec | comodulogram: ozkurt
[.......................... ] 67% | 34.85 sec | comodulogram: ozkurt
[........................... ] 68% | 35.20 sec | comodulogram: ozkurt
[............................ ] 70% | 35.58 sec | comodulogram: ozkurt
[............................ ] 72% | 35.90 sec | comodulogram: ozkurt
[............................. ] 73% | 36.26 sec | comodulogram: ozkurt
[.............................. ] 75% | 36.60 sec | comodulogram: ozkurt
[.............................. ] 77% | 37.20 sec | comodulogram: ozkurt
[............................... ] 78% | 37.73 sec | comodulogram: ozkurt
[................................ ] 80% | 38.67 sec | comodulogram: ozkurt
[................................ ] 82% | 39.54 sec | comodulogram: ozkurt
[................................. ] 83% | 40.14 sec | comodulogram: ozkurt
[.................................. ] 85% | 40.86 sec | comodulogram: ozkurt
[.................................. ] 87% | 41.35 sec | comodulogram: ozkurt
[................................... ] 88% | 41.92 sec | comodulogram: ozkurt
[.................................... ] 90% | 42.73 sec | comodulogram: ozkurt
[.................................... ] 92% | 43.46 sec | comodulogram: ozkurt
[..................................... ] 93% | 44.04 sec | comodulogram: ozkurt
[...................................... ] 95% | 44.66 sec | comodulogram: ozkurt
[...................................... ] 97% | 45.22 sec | comodulogram: ozkurt
[....................................... ] 98% | 46.41 sec | comodulogram: ozkurt
[........................................] 100% | 47.01 sec | comodulogram: ozkurt
[ ] 0% | 0.00 sec | comodulogram: penny
[ ] 2% | 2.27 sec | comodulogram: penny
[. ] 3% | 3.12 sec | comodulogram: penny
[.. ] 5% | 3.77 sec | comodulogram: penny
[.. ] 7% | 4.65 sec | comodulogram: penny
[... ] 8% | 5.58 sec | comodulogram: penny
[.... ] 10% | 6.36 sec | comodulogram: penny
[.... ] 12% | 7.07 sec | comodulogram: penny
[..... ] 13% | 8.12 sec | comodulogram: penny
[...... ] 15% | 9.21 sec | comodulogram: penny
[...... ] 17% | 10.47 sec | comodulogram: penny
[....... ] 18% | 11.77 sec | comodulogram: penny
[........ ] 20% | 12.68 sec | comodulogram: penny
[........ ] 22% | 13.48 sec | comodulogram: penny
[......... ] 23% | 14.27 sec | comodulogram: penny
[.......... ] 25% | 15.02 sec | comodulogram: penny
[.......... ] 27% | 15.74 sec | comodulogram: penny
[........... ] 28% | 16.53 sec | comodulogram: penny
[............ ] 30% | 17.21 sec | comodulogram: penny
[............ ] 32% | 18.01 sec | comodulogram: penny
[............. ] 33% | 18.81 sec | comodulogram: penny
[.............. ] 35% | 19.61 sec | comodulogram: penny
[.............. ] 37% | 20.45 sec | comodulogram: penny
[............... ] 38% | 21.26 sec | comodulogram: penny
[................ ] 40% | 22.14 sec | comodulogram: penny
[................ ] 42% | 22.95 sec | comodulogram: penny
[................. ] 43% | 24.00 sec | comodulogram: penny
[.................. ] 45% | 24.77 sec | comodulogram: penny
[.................. ] 47% | 25.49 sec | comodulogram: penny
[................... ] 48% | 26.24 sec | comodulogram: penny
[.................... ] 50% | 27.00 sec | comodulogram: penny
[.................... ] 52% | 27.76 sec | comodulogram: penny
[..................... ] 53% | 28.60 sec | comodulogram: penny
[...................... ] 55% | 29.37 sec | comodulogram: penny
[...................... ] 57% | 30.22 sec | comodulogram: penny
[....................... ] 58% | 30.99 sec | comodulogram: penny
[........................ ] 60% | 31.69 sec | comodulogram: penny
[........................ ] 62% | 32.40 sec | comodulogram: penny
[......................... ] 63% | 33.17 sec | comodulogram: penny
[.......................... ] 65% | 33.87 sec | comodulogram: penny
[.......................... ] 67% | 34.55 sec | comodulogram: penny
[........................... ] 68% | 35.33 sec | comodulogram: penny
[............................ ] 70% | 36.05 sec | comodulogram: penny
[............................ ] 72% | 36.86 sec | comodulogram: penny
[............................. ] 73% | 37.58 sec | comodulogram: penny
[.............................. ] 75% | 38.35 sec | comodulogram: penny
[.............................. ] 77% | 39.19 sec | comodulogram: penny
[............................... ] 78% | 40.04 sec | comodulogram: penny
[................................ ] 80% | 40.87 sec | comodulogram: penny
[................................ ] 82% | 41.78 sec | comodulogram: penny
[................................. ] 83% | 42.57 sec | comodulogram: penny
[.................................. ] 85% | 43.97 sec | comodulogram: penny
[.................................. ] 87% | 45.37 sec | comodulogram: penny
[................................... ] 88% | 46.60 sec | comodulogram: penny
[.................................... ] 90% | 47.84 sec | comodulogram: penny
[.................................... ] 92% | 49.20 sec | comodulogram: penny
[..................................... ] 93% | 50.54 sec | comodulogram: penny
[...................................... ] 95% | 51.87 sec | comodulogram: penny
[...................................... ] 97% | 53.21 sec | comodulogram: penny
[....................................... ] 98% | 54.60 sec | comodulogram: penny
[........................................] 100% | 55.68 sec | comodulogram: penny
[ ] 0% | 0.00 sec | comodulogram: tort
[ ] 2% | 2.58 sec | comodulogram: tort
[. ] 3% | 3.58 sec | comodulogram: tort
[.. ] 5% | 4.67 sec | comodulogram: tort
[.. ] 7% | 6.27 sec | comodulogram: tort
[... ] 8% | 8.03 sec | comodulogram: tort
[.... ] 10% | 9.90 sec | comodulogram: tort
[.... ] 12% | 11.70 sec | comodulogram: tort
[..... ] 13% | 13.50 sec | comodulogram: tort
[...... ] 15% | 15.24 sec | comodulogram: tort
[...... ] 17% | 16.95 sec | comodulogram: tort
[....... ] 18% | 18.92 sec | comodulogram: tort
[........ ] 20% | 20.51 sec | comodulogram: tort
[........ ] 22% | 22.48 sec | comodulogram: tort
[......... ] 23% | 23.81 sec | comodulogram: tort
[.......... ] 25% | 25.24 sec | comodulogram: tort
[.......... ] 27% | 26.52 sec | comodulogram: tort
[........... ] 28% | 28.08 sec | comodulogram: tort
[............ ] 30% | 29.68 sec | comodulogram: tort
[............ ] 32% | 31.39 sec | comodulogram: tort
[............. ] 33% | 33.40 sec | comodulogram: tort
[.............. ] 35% | 35.24 sec | comodulogram: tort
[.............. ] 37% | 37.03 sec | comodulogram: tort
[............... ] 38% | 38.90 sec | comodulogram: tort
[................ ] 40% | 40.87 sec | comodulogram: tort
[................ ] 42% | 42.95 sec | comodulogram: tort
[................. ] 43% | 45.10 sec | comodulogram: tort
[.................. ] 45% | 47.12 sec | comodulogram: tort
[.................. ] 47% | 48.99 sec | comodulogram: tort
[................... ] 48% | 51.03 sec | comodulogram: tort
[.................... ] 50% | 52.87 sec | comodulogram: tort
[.................... ] 52% | 54.69 sec | comodulogram: tort
[..................... ] 53% | 56.80 sec | comodulogram: tort
[...................... ] 55% | 58.72 sec | comodulogram: tort
[...................... ] 57% | 60.65 sec | comodulogram: tort
[....................... ] 58% | 62.69 sec | comodulogram: tort
[........................ ] 60% | 64.75 sec | comodulogram: tort
[........................ ] 62% | 66.72 sec | comodulogram: tort
[......................... ] 63% | 68.68 sec | comodulogram: tort
[.......................... ] 65% | 70.63 sec | comodulogram: tort
[.......................... ] 67% | 72.58 sec | comodulogram: tort
[........................... ] 68% | 74.84 sec | comodulogram: tort
[............................ ] 70% | 76.63 sec | comodulogram: tort
[............................ ] 72% | 78.80 sec | comodulogram: tort
[............................. ] 73% | 81.34 sec | comodulogram: tort
[.............................. ] 75% | 83.12 sec | comodulogram: tort
[.............................. ] 77% | 84.51 sec | comodulogram: tort
[............................... ] 78% | 86.28 sec | comodulogram: tort
[................................ ] 80% | 87.57 sec | comodulogram: tort
[................................ ] 82% | 88.95 sec | comodulogram: tort
[................................. ] 83% | 90.20 sec | comodulogram: tort
[.................................. ] 85% | 91.46 sec | comodulogram: tort
[.................................. ] 87% | 93.09 sec | comodulogram: tort
[................................... ] 88% | 94.34 sec | comodulogram: tort
[.................................... ] 90% | 95.77 sec | comodulogram: tort
[.................................... ] 92% | 97.54 sec | comodulogram: tort
[..................................... ] 93% | 98.80 sec | comodulogram: tort
[...................................... ] 95% | 100.35 sec | comodulogram: tort
[...................................... ] 97% | 101.63 sec | comodulogram: tort
[....................................... ] 98% | 103.23 sec | comodulogram: tort
[........................................] 100% | 104.74 sec | comodulogram: tort
[ ] 0% | 0.00 sec | comodulogram: DAR(10, 1)
[ ] 2% | 0.07 sec | comodulogram: DAR(10, 1)
[. ] 3% | 0.09 sec | comodulogram: DAR(10, 1)
[.. ] 5% | 0.10 sec | comodulogram: DAR(10, 1)
[.. ] 7% | 0.11 sec | comodulogram: DAR(10, 1)
[... ] 8% | 0.13 sec | comodulogram: DAR(10, 1)
[.... ] 10% | 0.14 sec | comodulogram: DAR(10, 1)
[.... ] 12% | 0.16 sec | comodulogram: DAR(10, 1)
[..... ] 13% | 0.18 sec | comodulogram: DAR(10, 1)
[...... ] 15% | 0.19 sec | comodulogram: DAR(10, 1)
[...... ] 17% | 0.21 sec | comodulogram: DAR(10, 1)
[....... ] 18% | 0.23 sec | comodulogram: DAR(10, 1)
[........ ] 20% | 0.28 sec | comodulogram: DAR(10, 1)
[........ ] 22% | 0.30 sec | comodulogram: DAR(10, 1)
[......... ] 23% | 0.47 sec | comodulogram: DAR(10, 1)
[.......... ] 25% | 0.58 sec | comodulogram: DAR(10, 1)
[.......... ] 27% | 0.66 sec | comodulogram: DAR(10, 1)
[........... ] 28% | 0.76 sec | comodulogram: DAR(10, 1)
[............ ] 30% | 0.84 sec | comodulogram: DAR(10, 1)
[............ ] 32% | 0.92 sec | comodulogram: DAR(10, 1)
[............. ] 33% | 1.00 sec | comodulogram: DAR(10, 1)
[.............. ] 35% | 1.11 sec | comodulogram: DAR(10, 1)
[.............. ] 37% | 5.54 sec | comodulogram: DAR(10, 1)
[............... ] 38% | 5.61 sec | comodulogram: DAR(10, 1)
[................ ] 40% | 5.75 sec | comodulogram: DAR(10, 1)
[................ ] 42% | 5.88 sec | comodulogram: DAR(10, 1)
[................. ] 43% | 6.03 sec | comodulogram: DAR(10, 1)
[.................. ] 45% | 6.17 sec | comodulogram: DAR(10, 1)
[.................. ] 47% | 6.34 sec | comodulogram: DAR(10, 1)
[................... ] 48% | 6.50 sec | comodulogram: DAR(10, 1)
[.................... ] 50% | 6.63 sec | comodulogram: DAR(10, 1)
[.................... ] 52% | 6.77 sec | comodulogram: DAR(10, 1)
[..................... ] 53% | 6.92 sec | comodulogram: DAR(10, 1)
[...................... ] 55% | 11.76 sec | comodulogram: DAR(10, 1)
[...................... ] 57% | 11.83 sec | comodulogram: DAR(10, 1)
[....................... ] 58% | 11.97 sec | comodulogram: DAR(10, 1)
[........................ ] 60% | 12.12 sec | comodulogram: DAR(10, 1)
[........................ ] 62% | 12.25 sec | comodulogram: DAR(10, 1)
[......................... ] 63% | 12.39 sec | comodulogram: DAR(10, 1)
[.......................... ] 65% | 12.53 sec | comodulogram: DAR(10, 1)
[.......................... ] 67% | 12.70 sec | comodulogram: DAR(10, 1)
[........................... ] 68% | 12.83 sec | comodulogram: DAR(10, 1)
[............................ ] 70% | 12.90 sec | comodulogram: DAR(10, 1)
[............................ ] 72% | 12.94 sec | comodulogram: DAR(10, 1)
[............................. ] 73% | 15.79 sec | comodulogram: DAR(10, 1)
[.............................. ] 75% | 15.83 sec | comodulogram: DAR(10, 1)
[.............................. ] 77% | 15.90 sec | comodulogram: DAR(10, 1)
[............................... ] 78% | 15.96 sec | comodulogram: DAR(10, 1)
[................................ ] 80% | 16.03 sec | comodulogram: DAR(10, 1)
[................................ ] 82% | 16.13 sec | comodulogram: DAR(10, 1)
[................................. ] 83% | 16.20 sec | comodulogram: DAR(10, 1)
[.................................. ] 85% | 16.26 sec | comodulogram: DAR(10, 1)
[.................................. ] 87% | 16.30 sec | comodulogram: DAR(10, 1)
[................................... ] 88% | 16.38 sec | comodulogram: DAR(10, 1)
[.................................... ] 90% | 16.46 sec | comodulogram: DAR(10, 1)
[.................................... ] 92% | 19.70 sec | comodulogram: DAR(10, 1)
[..................................... ] 93% | 19.72 sec | comodulogram: DAR(10, 1)
[...................................... ] 95% | 19.78 sec | comodulogram: DAR(10, 1)
[...................................... ] 97% | 19.81 sec | comodulogram: DAR(10, 1)
[....................................... ] 98% | 19.88 sec | comodulogram: DAR(10, 1)
[........................................] 100% | 19.95 sec | comodulogram: DAR(10, 1)
[........................................] 100% | 25.40 sec | comodulogram: DAR(10, 1) /home/tom/work/github/pactools/examples/plot_spurious_pac.py:118: UserWarning: Matplotlib is currently using agg, which is a non-GUI backend, so cannot show the figure.
plt.show()
Total running time of the script: ( 3 minutes 54.364 seconds)